5.10 Teste de Hipóteses
- Seção 3.4 de (Paulino, Turkman, and Murteira 2003)
- Capítulo 9 de (S. J. Press 2003)
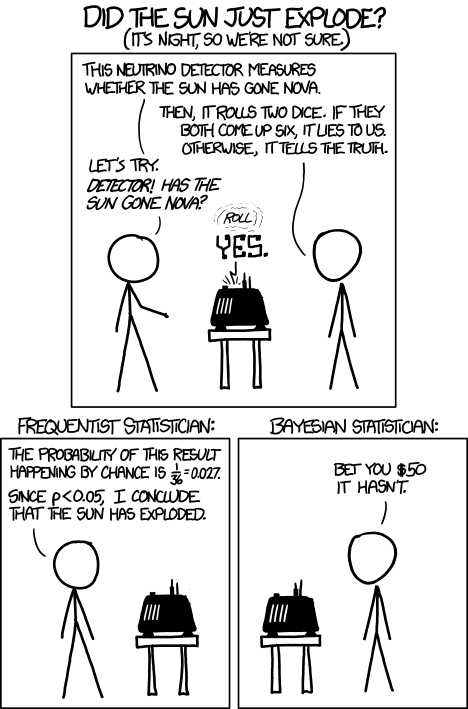
“Did the sun just explode?” by xkcd.com
5.10.1 Fator de Bayes
- (Kass and Raftery 1995)
- Seção 3.4.1 de (Paulino, Turkman, and Murteira 2003)
- Seção 9.5.1 de (S. J. Press 2003)
5.10.1.1 Pacote BayesFactor
(Morey and Rouder 2022) apresentam o pacote BayesFactor.
Exemplo 5.13 Exemplo da documentação.

## classical paired t test
t.test(x = sleep2$extra[sleep2$group==1],
y = sleep2$extra[sleep2$group==2], paired = TRUE)##
## Paired t-test
##
## data: sleep2$extra[sleep2$group == 1] and sleep2$extra[sleep2$group == 2]
## t = -4.0621, df = 9, p-value = 0.002833
## alternative hypothesis: true mean difference is not equal to 0
## 95 percent confidence interval:
## -2.4598858 -0.7001142
## sample estimates:
## mean difference
## -1.58# classical Wilcoxon (signed rank test (median = 0 versus median > 0)
wilcox.test(x = sleep2$extra[sleep2$group==1],
y = sleep2$extra[sleep2$group==2], paired = TRUE)##
## Wilcoxon signed rank test with continuity correction
##
## data: sleep2$extra[sleep2$group == 1] and sleep2$extra[sleep2$group == 2]
## V = 0, p-value = 0.009091
## alternative hypothesis: true location shift is not equal to 0## bayesian paired t test
ttestBF(x = sleep2$extra[sleep2$group == 1],
y = sleep2$extra[sleep2$group == 2], paired = TRUE)## Bayes factor analysis
## --------------
## [1] Alt., r=0.707 : 17.25888 ±0%
##
## Against denominator:
## Null, mu = 0
## ---
## Bayes factor type: BFoneSample, JZSExercício 5.8 Veja a documentação de pingouin.bayesfactor_ttest no link a seguir.
https://pingouin-stats.org/build/html/generated/pingouin.bayesfactor_ttest.html#pingouin.bayesfactor_ttest
Exemplo 5.14 Exemplo Moon and Agression.
# lendo dados
ma <- read.csv('https://filipezabala.com/data/moon-aggression.csv')
# gráfico
boxplot(ma)
##
## Paired t-test
##
## data: ma$Moon and ma$Other
## t = 6.4518, df = 14, p-value = 1.518e-05
## alternative hypothesis: true mean difference is not equal to 0
## 95 percent confidence interval:
## 1.623968 3.241365
## sample estimates:
## mean difference
## 2.432667# classical Wilcoxon (signed rank test (median = 0 versus median > 0)
wilcox.test(x = ma$Moon,
y = ma$Other, paired = TRUE)##
## Wilcoxon signed rank exact test
##
## data: ma$Moon and ma$Other
## V = 119, p-value = 0.0001221
## alternative hypothesis: true location shift is not equal to 0## Bayes factor analysis
## --------------
## [1] Alt., r=0.707 : 1521.194 ±0%
##
## Against denominator:
## Null, mu = 0
## ---
## Bayes factor type: BFoneSample, JZS5.10.1.2 Pacote BFpack
(Mulder et al. 2021) apresentam o pacote BFpack.
Exercício 5.9 Veja a vinheta de BFpack em https://cran.r-project.org/package=BFpack.
5.10.2 FBST
FBST é o acrônimo para Full Bayesian Significance Test.
- Proposta de (Carlos Alberto de Bragança Pereira and Stern 1999) para testar hipóteses precisas (sharp hypotheses)
- Amplamente revisado em (Carlos A. de Bragança Pereira et al. 2008) e (Carlos Alberto de B. Pereira and Stern 2020)
- https://jreduardo.github.io/ce227-bayes/04-fbst.html
5.10.2.1 Pacote fbst
(Kelter 2020) apresenta o pacote de R fbst.
# https://cran.r-project.org/package=fbst
library(fbst)
set.seed(57)
grp1 <- rnorm(50,0,1.5)
grp2 <- rnorm(50,0.8,3.2)
p <- as.vector(BayesFactor::ttestBF(x=grp1,y=grp2,
posterior = TRUE, iterations = 3000,
rscale = "medium")[,4])
# flat reference function
res <- fbst(posteriorDensityDraws = p, nullHypothesisValue = 0,
dimensionTheta = 2, dimensionNullset = 1)
summary(res)## Full Bayesian Significance Test for testing a sharp hypothesis against its alternative:
## Reference function: Flat
## Hypothesis H_0:Parameter= 0 against its alternative H_1
## Bayesian e-value against H_0: 0.92749
## Standardized e-value: 0.02197123
# medium Cauchy C(0,1) reference function
res_med <- fbst(posteriorDensityDraws = p, nullHypothesisValue = 0,
dimensionTheta = 2, dimensionNullset = 1, FUN = dcauchy,
par = list(location = 0, scale = sqrt(2)/2))
summary(res_med)## Full Bayesian Significance Test for testing a sharp hypothesis against its alternative:
## Reference function: User-defined
## Hypothesis H_0:Parameter= 0 against its alternative H_1
## Bayesian e-value against H_0: 0.9538276
## Standardized e-value: 0.01313566
5.10.3 Valores-p bayesiano
O valor-p preditivo a posteriori (Paulino, Turkman, and Murteira 2003) e a probabilidade de direção (Makowski et al. 2019) são reportados na literatura como ‘valores-pr bayesianos’.
5.10.4 Combinando ferramentas
(Gannon, Pereira, and Polpo 2019)
5.10.4.1 Comparando proporções
The observed results in this example are that only one of the patients in the control arm responded positively, but in the case arm there were four positive outcomes. (Gannon, Pereira, and Polpo 2019, 217)
tab <- matrix(c(1,7, 4,4), nrow = 2, byrow = TRUE)
rownames(tab) <- c('C', 'T') # Controle/Tratamento
colnames(tab) <- c('+', '-') # Respondeu positivamente/negativamente
(tab <- as.table(tab))## + -
## C 1 7
## T 4 4##
## Pearson's Chi-squared test
##
## data: tab
## X-squared = 2.6182, df = 1, p-value = 0.1056##
## Pearson's Chi-squared test with Yates' continuity correction
##
## data: tab
## X-squared = 1.1636, df = 1, p-value = 0.2807##
## Fisher's Exact Test for Count Data
##
## data: tab
## p-value = 0.2821
## alternative hypothesis: true odds ratio is not equal to 1
## 95 percent confidence interval:
## 0.002553456 2.416009239
## sample estimates:
## odds ratio
## 0.1624254# Fator de Bayes
fh <- function(x,y){
(choose(8,x)*choose(8,y))/(17*choose(16,x+y))
}
fa <- 1/81
bf <- function(x,y){
fh(x,y)/fa
}
BF <- matrix(nrow = 9, ncol = 9)
rownames(BF) <- paste0('x.',0:8)
colnames(BF) <- paste0('y.',0:8)
for(i in 0:8){
for(j in 0:8){
BF[i+1,j+1] <- bf(j,i)
}
}
addmargins(round(BF,3)) # note a questão do arredondamento## y.0 y.1 y.2 y.3 y.4 y.5 y.6 y.7 y.8 Sum
## x.0 4.765 2.382 1.112 0.476 0.183 0.061 0.017 0.003 0.000 8.999
## x.1 2.382 2.541 1.906 1.173 0.611 0.267 0.093 0.024 0.003 9.000
## x.2 1.112 1.906 2.052 1.710 1.166 0.653 0.290 0.093 0.017 8.999
## x.3 0.476 1.173 1.710 1.866 1.633 1.161 0.653 0.267 0.061 9.000
## x.4 0.183 0.611 1.166 1.633 1.814 1.633 1.166 0.611 0.183 9.000
## x.5 0.061 0.267 0.653 1.161 1.633 1.866 1.710 1.173 0.476 9.000
## x.6 0.017 0.093 0.290 0.653 1.166 1.710 2.052 1.906 1.112 8.999
## x.7 0.003 0.024 0.093 0.267 0.611 1.173 1.906 2.541 2.382 9.000
## x.8 0.000 0.003 0.017 0.061 0.183 0.476 1.112 2.382 4.765 8.999
## Sum 8.999 9.000 8.999 9.000 9.000 9.000 8.999 9.000 8.999 80.996## [1] 0.6108597## [1] 0.09228027