Capítulo 5
Solução. ??
lambda <- 1 # valores iniciais
Nsim <- 5000 # número de simulações
X <- rep(runif(1,0,10), Nsim) # inicia a cadeia
Naccept <- 0
for (i in 2:Nsim){
Y <- runif(1,0,10)
rho <- min((dexp(Y,lambda)/dexp(X[i-1],lambda)) * (10/10), 1)
accept <- runif(1) < rho
X[i] <- X[i-1] + (Y - X[i-1])*accept
Naccept <- Naccept+accept
}
Naccept/Nsim # taxa de aceitação## [1] 0.1916Validando.
ex <- rexp(Nsim,lambda) # exponencial via stats::rexp
ks.test(jitter(X), ex) # teste de aderência de Kolmogorov-Smirnov##
## Asymptotic two-sample Kolmogorov-Smirnov test
##
## data: jitter(X) and ex
## D = 0.0752, p-value = 1.05e-12
## alternative hypothesis: two-sidedplot(ecdf(X), col = 'blue', main = 'Função de distribuição acumulada empírica')
plot(ecdf(ex), col = 'red', add = TRUE)
legend(4, .9, legend=c('Metropolis-Hastings', 'stats::rexp'), lty = 1,
col=c("red", "blue"), cex=0.8, bty = 'n')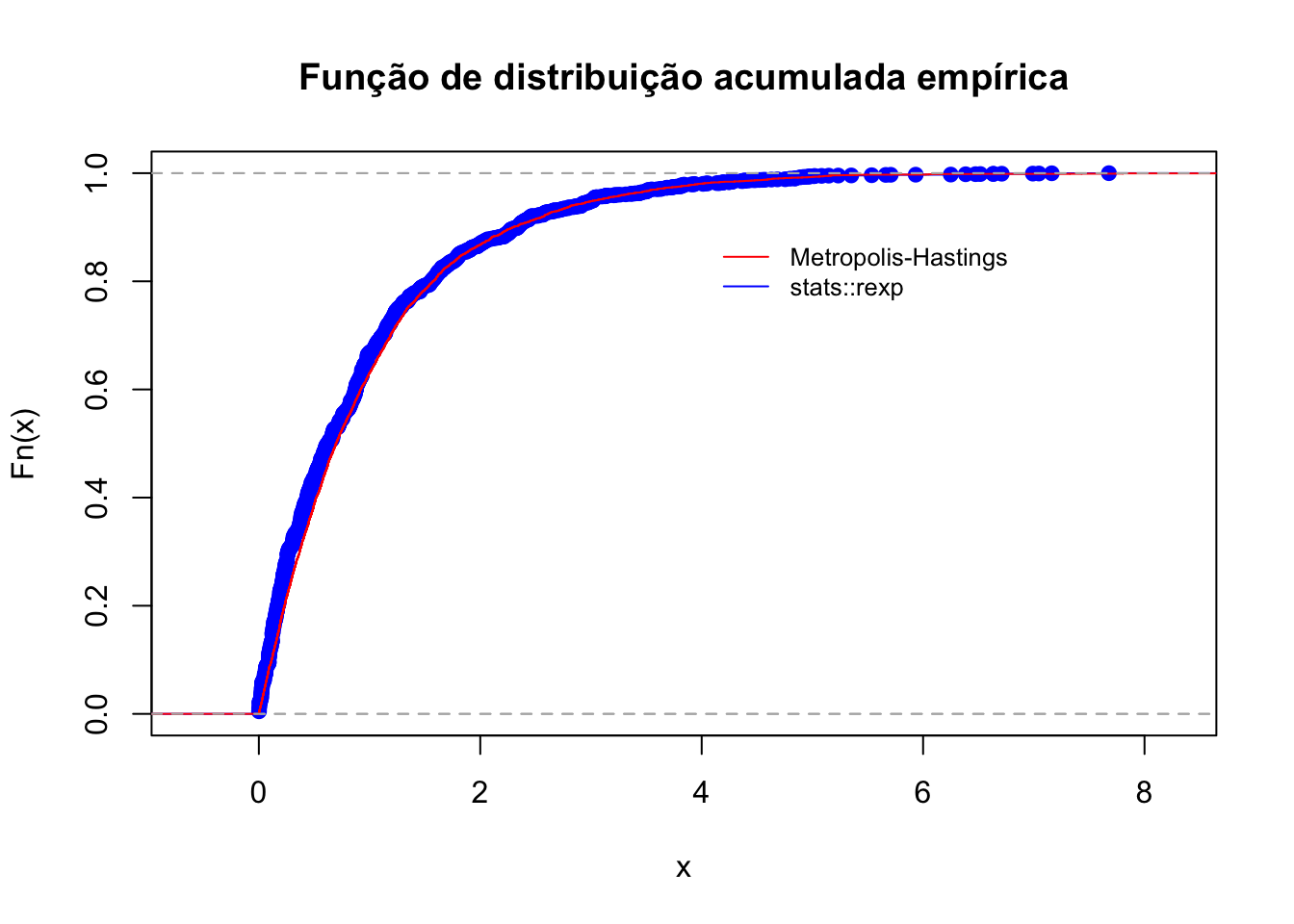
## [1] 1## [1] 0.9523171## [1] 1## [1] 1.036261par(mfrow=c(1,2))
hist(X, 30, freq = FALSE, main = 'Exp(1) via M-H')
curve(dexp(x,lambda), col = 'blue', add = TRUE)
hist(ex, 30, freq = FALSE, main = 'Exp(1) via stats::rexp')
curve(dexp(x,lambda), col = 'red', add = TRUE)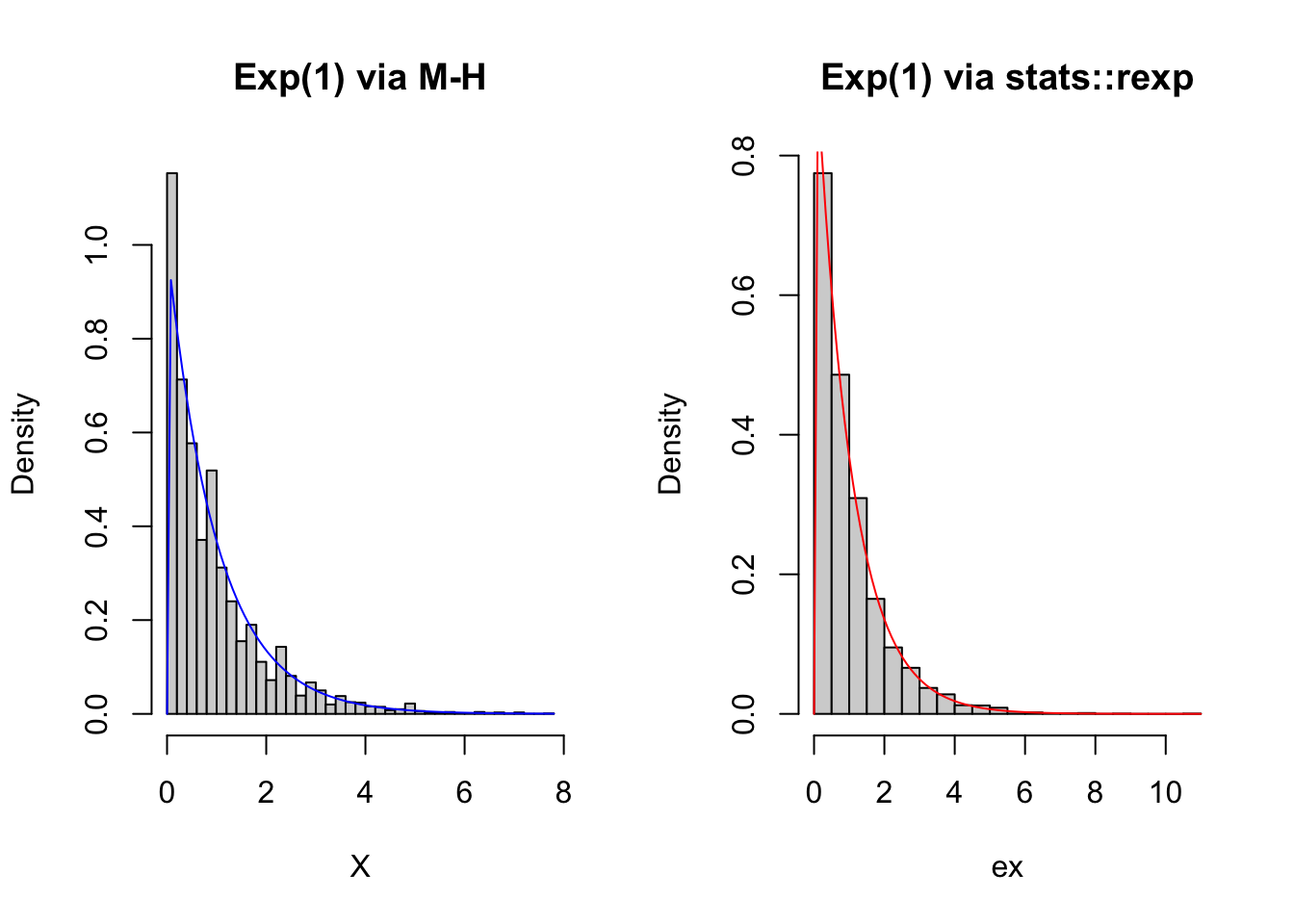
Solução. ??
- Teoria da Informação. Como você quantifica e mede a informação. Veja https://www.youtube.com/watch?v=4toWtb7PRH4#t=3m10s.
- Informação. \(I = -\log p(X)\), \(X\) evento. Maior a probabilidade|dados, menor a informação. Veja https://www.youtube.com/watch?v=4toWtb7PRH4#t=3m58s.
- Entropia. É a informação média de uma V.A. Discreta. \(H = - \sum_{i=1}^{|R_x|} p(X_i) \log p(X_i)\). Veja https://www.youtube.com/watch?v=4toWtb7PRH4#t=10m08s.
- Entropia diferencial. É a informação média de uma V.A. Contínua. \(H = - \int_{i=1}^{|R_x|} p(X_i) \log p(X_i)\). Veja https://www.youtube.com/watch?v=4toWtb7PRH4#t=17m23s. Resumo do vídeo 1, https://www.youtube.com/watch?v=4toWtb7PRH4#t=22m28s.
- Divergência de Kullback–Leibler. Entropia relativa. Mede a distância entre duas distribuições de probabilidade. \(KL(p||q)\): divergência de KL em relação a \(p\) contra \(q\). Veja https://www.youtube.com/watch?v=uKxtmkfeuxg#t=1m09s.
\(KL(p||q) = - \sum p(x) \log q(x) - (- \sum p(x) \log p(x))\)
\(KL(p||q) = \sum p(x) \log p(x) - \sum p(x) \log q(x)\)
\(KL(p||q) = \sum p(x) (\log p(x) - \log q(x))\)
\(KL(p||q) = \sum p(x) \log (p(x)/q(x))\)
\(KL(p||q) = - \sum p(x) \log (q(x)/p(x))\)
\(KL(q||p) = - \sum q(x) \log p(x) - (- \sum q(x) \log q(x))\)
- \(KL \ge 0\).
- KL não é simétrica.
Usando \(q(z)\) para estimar \(p(z|x)\), pode-se utilizar KL para medir a qualidade da estimação \(L = \sum q(z) \log (p(x,z)/q(x))\) # Lower bound
\(KL(q(z)||p(z|x)) = - \sum q(z) \log (p(z|x)/q(z))\)
\(KL + L = \log p(X)\)
≥0 <0 <0
\(\log p(X)\) é constante, logo maximizar L é o mesmo que minimizar KL.
- Inferência Variacional. Objetivo: obter uma distribuição condicional a partir de uma conjunta. https://www.youtube.com/watch?v=BxkZcS1pLpw#t=3m28s