11.12 Modelos ARCH
11.12.1 ARCH
(R. F. Engle 1982) propõe o modelo ARCH (Autoregressive Conditional Heteroscedasticity), que permite modelar a variância condicional de uma série em função dos quadrados dos retornos passados. A variância inconstante ao longo do tempo é chamada heterocedasticidade.
O modelo ARCH(\(s\)) pode ser expresso por
\[\begin{equation} y_t = a_t \tag{11.35} \end{equation}\]
\[\begin{equation} a_t = \varepsilon_t \sqrt{h_t} \tag{11.36} \end{equation}\]
\[\begin{equation} h_t = \alpha_0 + \sum_{i=1}^s \alpha_i y_{t-i}^2 \tag{11.37} \end{equation}\]
\[\begin{equation} \varepsilon_t \sim WN(0,1) \tag{11.38} \end{equation}\]
Demonstra-se que \(a_t \sim WN(0,\sigma^2_a)\), i.e., \(a_t\) tem variância constante tal que
\[\begin{equation} \sigma^2_a = \frac{\alpha_0}{1-\sum_{i=1}^s \alpha_i} \tag{11.39} \end{equation}\]
Portanto \(a_t\) possui estacionariedade fraca ou de segunda ordem. Para mais detalhes recomenda-se (R. F. Engle 1982), (Shephard 1996) e (G. E. P. Box, Jenkins, and Reinsel 2008, 413).
11.12.1.1 Clássico
Exemplo 11.54 Adaptado das documentações de tseries::garch() (Trapletti and Hornik 2024) e rugarch::ugarchspec() (Galanos 2025), simulando e ajustando um modelo ARCH\((2)\).
# libs
library(tseries)
library(rugarch)
# número de observações simuladas, 100 primeiras descartadas
n <- 1100
# coeficientes ARCH(2): alfa_0=0.1, alfa_1=0.5 e alfa_2=0.2
a <- c(0.1, 0.5, 0.2)
# \varepsilon_t, Eq. 11.38
set.seed(42); e <- rnorm(n)
# vetor para conter a série temporal yt simulada (double-precision vector)
y <- double(n)
# \sigma_a^2, Eq. 11.39
sigma2a <- sqrt(a[1]/(1-a[2]-a[3]))
# 2 primeiros valores de yt, Eq. 11.35
set.seed(314); y[1:2] <- rnorm(2, sd = sigma2a)
# gera processo ARCH(2)
for(i in 3:n){
# Eq. 11.37
ht <- a[1] + a[2]*y[i-1]^2 + a[3]*y[i-2]^2
# Eqs. 11.35 e 11.36
y[i] <- e[i]*sqrt(ht)
}
# descartando as 100 primeiras observações
y <- ts(y[101:n])
# ARCH(2)
spec <- ugarchspec(variance.model = list(garchOrder = c(2,0)),
mean.model = list(armaOrder = c(0,0)))
fit_arch <- ugarchfit(spec, y)
# diagnóstico
fit_arch##
## *---------------------------------*
## * GARCH Model Fit *
## *---------------------------------*
##
## Conditional Variance Dynamics
## -----------------------------------
## GARCH Model : sGARCH(2,0)
## Mean Model : ARFIMA(0,0,0)
## Distribution : norm
##
## Optimal Parameters
## ------------------------------------
## Estimate Std. Error t value Pr(>|t|)
## mu -0.015908 0.012160 -1.3082 0.190815
## omega 0.095962 0.009116 10.5266 0.000000
## alpha1 0.500586 0.063300 7.9082 0.000000
## alpha2 0.225688 0.049399 4.5686 0.000005
##
## Robust Standard Errors:
## Estimate Std. Error t value Pr(>|t|)
## mu -0.015908 0.011578 -1.3739 0.169471
## omega 0.095962 0.009047 10.6065 0.000000
## alpha1 0.500586 0.063524 7.8802 0.000000
## alpha2 0.225688 0.057314 3.9378 0.000082
##
## LogLikelihood : -661.2625
##
## Information Criteria
## ------------------------------------
##
## Akaike 1.3305
## Bayes 1.3502
## Shibata 1.3305
## Hannan-Quinn 1.3380
##
## Weighted Ljung-Box Test on Standardized Residuals
## ------------------------------------
## statistic p-value
## Lag[1] 0.06124 0.8045
## Lag[2*(p+q)+(p+q)-1][2] 0.16723 0.8747
## Lag[4*(p+q)+(p+q)-1][5] 1.17259 0.8195
## d.o.f=0
## H0 : No serial correlation
##
## Weighted Ljung-Box Test on Standardized Squared Residuals
## ------------------------------------
## statistic p-value
## Lag[1] 0.197 0.6571
## Lag[2*(p+q)+(p+q)-1][5] 3.104 0.3884
## Lag[4*(p+q)+(p+q)-1][9] 6.582 0.2367
## d.o.f=2
##
## Weighted ARCH LM Tests
## ------------------------------------
## Statistic Shape Scale P-Value
## ARCH Lag[3] 0.1861 0.500 2.000 0.6662
## ARCH Lag[5] 1.9632 1.440 1.667 0.4794
## ARCH Lag[7] 3.8599 2.315 1.543 0.3672
##
## Nyblom stability test
## ------------------------------------
## Joint Statistic: 0.57
## Individual Statistics:
## mu 0.02503
## omega 0.35414
## alpha1 0.25327
## alpha2 0.21252
##
## Asymptotic Critical Values (10% 5% 1%)
## Joint Statistic: 1.07 1.24 1.6
## Individual Statistic: 0.35 0.47 0.75
##
## Sign Bias Test
## ------------------------------------
## t-value prob sig
## Sign Bias 1.1810 0.2379
## Negative Sign Bias 0.9791 0.3278
## Positive Sign Bias 0.3806 0.7036
## Joint Effect 1.5812 0.6637
##
##
## Adjusted Pearson Goodness-of-Fit Test:
## ------------------------------------
## group statistic p-value(g-1)
## 1 20 16.00 0.6573
## 2 30 30.98 0.3664
## 3 40 37.68 0.5300
## 4 50 47.80 0.5218
##
##
## Elapsed time : 0.06077385# previsão 20 períodos à frente
fcast <- ugarchforecast(fit_arch, n.ahead = 20)
plot(fcast, which = 1) # 1 = Conditional mean forecast
11.12.2 GARCH
(Bollerslev 1986) estendeu o trabalho original de Engle, propondo o modelo GARCH (Generalized Autoregressive Conditional Heteroscedasticity), que permite que a variância condicional não dependa somente dos quadrados dos retornos passados, mas também da própria variância passada. O modelo GARCH\((r,s)\) é definido pelas Eq. (11.35), (11.36) e (11.38), com \(h_t\) dado pela Eq. (11.40).
\[\begin{equation} h_t = \alpha_0 + \sum_{i=1}^s \alpha_i y_{t-i}^2 + \sum_{i=1}^r \beta_i h_{t-i} \tag{11.40} \end{equation}\]
Para esta formulação \(a_t\) também possui variância constante, definida pela Eq. (11.41).
\[\begin{equation} \sigma^2_a = \frac{\alpha_0}{1-\sum_{i=1}^s \alpha_i - \sum_{i=1}^r \beta_i} \tag{11.41} \end{equation}\]
11.12.2.1 Clássico
Exemplo 11.55 Adaptado da documentação de tseries::garch(), simulando e ajustando um modelo GARCH(1,1). Lembre-se que “[t]he [a]utocovariances [d]o [n]ot [a]lways [d]ecrease” (Francq and Zakoian 2010, 51).
# libs
library(tseries)
library(rugarch)
# número de observações simuladas, 100 primeiras descartadas
n <- 1100
# coeficientes ARCH(1): alfa_0=0.1 e alfa_1=0.5
a <- c(0.1, 0.5)
# coeficiente GARCH(s,1): beta_1=0.3
b <- c(0.3)
# \varepsilon_t, Eq. 11.38
set.seed(42); e <- rnorm(n)
# vetor para conter a série temporal yt simulada (double-precision vector)
y <- double(n)
# vetor para conter a série temporal de ht
h <- double(n)
# Eqs. 11.34 e 11.40
set.seed(314); y[1] <- rnorm(1, sd = sqrt(a[1]/(1-a[2]-b[1])))
# gera processo GARCH(1,1)
for(i in 2:n){
# Eq. 11.39
h[i] <- a[1] + a[2]*y[i-1]^2 + b[1]*h[i-1]
# Eqs. 11.34 e 11.35
y[i] <- e[i]*sqrt(h[i])
}
# descartando as 100 primeiras observações
y <- ts(y[101:n])
# GARCH(1,1)
spec <- ugarchspec(variance.model = list(garchOrder = c(1,1)),
mean.model = list(armaOrder = c(0,0)))
fit_garch <- ugarchfit(spec, y)
# diagnóstico
fit_garch##
## *---------------------------------*
## * GARCH Model Fit *
## *---------------------------------*
##
## Conditional Variance Dynamics
## -----------------------------------
## GARCH Model : sGARCH(1,1)
## Mean Model : ARFIMA(0,0,0)
## Distribution : norm
##
## Optimal Parameters
## ------------------------------------
## Estimate Std. Error t value Pr(>|t|)
## mu -0.018259 0.014908 -1.2248 0.22064
## omega 0.073548 0.014315 5.1377 0.00000
## alpha1 0.468977 0.061610 7.6121 0.00000
## beta1 0.397669 0.061347 6.4823 0.00000
##
## Robust Standard Errors:
## Estimate Std. Error t value Pr(>|t|)
## mu -0.018259 0.014069 -1.2978 0.194345
## omega 0.073548 0.017321 4.2462 0.000022
## alpha1 0.468977 0.070549 6.6475 0.000000
## beta1 0.397669 0.084187 4.7237 0.000002
##
## LogLikelihood : -866.4694
##
## Information Criteria
## ------------------------------------
##
## Akaike 1.7409
## Bayes 1.7606
## Shibata 1.7409
## Hannan-Quinn 1.7484
##
## Weighted Ljung-Box Test on Standardized Residuals
## ------------------------------------
## statistic p-value
## Lag[1] 0.05405 0.8162
## Lag[2*(p+q)+(p+q)-1][2] 0.17145 0.8721
## Lag[4*(p+q)+(p+q)-1][5] 1.21927 0.8084
## d.o.f=0
## H0 : No serial correlation
##
## Weighted Ljung-Box Test on Standardized Squared Residuals
## ------------------------------------
## statistic p-value
## Lag[1] 0.872 0.3504
## Lag[2*(p+q)+(p+q)-1][5] 4.705 0.1783
## Lag[4*(p+q)+(p+q)-1][9] 7.756 0.1437
## d.o.f=2
##
## Weighted ARCH LM Tests
## ------------------------------------
## Statistic Shape Scale P-Value
## ARCH Lag[3] 0.000114 0.500 2.000 0.9915
## ARCH Lag[5] 3.394671 1.440 1.667 0.2374
## ARCH Lag[7] 4.871650 2.315 1.543 0.2378
##
## Nyblom stability test
## ------------------------------------
## Joint Statistic: 0.4709
## Individual Statistics:
## mu 0.02895
## omega 0.22996
## alpha1 0.30454
## beta1 0.28857
##
## Asymptotic Critical Values (10% 5% 1%)
## Joint Statistic: 1.07 1.24 1.6
## Individual Statistic: 0.35 0.47 0.75
##
## Sign Bias Test
## ------------------------------------
## t-value prob sig
## Sign Bias 1.5441 0.1229
## Negative Sign Bias 1.0226 0.3067
## Positive Sign Bias 0.7825 0.4341
## Joint Effect 2.4511 0.4842
##
##
## Adjusted Pearson Goodness-of-Fit Test:
## ------------------------------------
## group statistic p-value(g-1)
## 1 20 18.20 0.5091
## 2 30 25.88 0.6319
## 3 40 45.84 0.2096
## 4 50 54.80 0.2640
##
##
## Elapsed time : 0.05178595# previsão 20 períodos à frente
fcast <- ugarchforecast(fit_garch, n.ahead = 20)
plot(fcast, which = 1) # 1 = Conditional mean forecast
11.12.2.2 Bayesiano
A função bayesforecast::garch() (Matamoros and Torres 2020) implementa um modelo GARCH\((s,k,h)\) baseado em (Ardia and Hoogerheide 2010) e (Fonseca et al. 2019). As três componentes \((s,k,h) \equiv (s,r,h)\) são a ordem do ARCH (\(s\)), a ordem do GARCH (\(k \equiv r\)) e a ordem do MGARCH (\(h\)) (Bollerslev, Engle, and Wooldridge 1988). Segundo a documentação, as prioris padrão utilizadas no modelo MGARCH são:
- ar ~ normal(0,0.5)
- ma ~ normal(0,0.5)
- mu0 ~ t-student(0,2.5,6)
- sigma0 ~ t-student(0,1,7)
- arch ~ normal(0,0.5)
- garch ~ normal(0,0.5)
- mgarch ~ normal(0,0.5)
- dfv ~ gamma(2,0.1)
- breg ~ t-student(0,2.5,6)
Para mais detalhes veja (Engle Robert and Kevin 2001) e (R. Engle 2002).
Exemplo 11.56 Considere a série \(y_t\) gerada no Exemplo 11.55.
# libs
library(bayesforecast)
# GARCH(1,1)
fit_garch_bayes <- bayesforecast::garch(y, order = c(1,1,0))
fit_garch_varstan <- bayesforecast::varstan(fit_garch_bayes, iter = 500,
chains = 1, quiet = TRUE)##
## SAMPLING FOR MODEL 'tgarch' NOW (CHAIN 1).
## Chain 1:
## Chain 1: Gradient evaluation took 0.001832 seconds
## Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 18.32 seconds.
## Chain 1: Adjust your expectations accordingly!
## Chain 1:
## Chain 1:
## Chain 1: Iteration: 1 / 500 [ 0%] (Warmup)
## Chain 1: Iteration: 50 / 500 [ 10%] (Warmup)
## Chain 1: Iteration: 100 / 500 [ 20%] (Warmup)
## Chain 1: Iteration: 150 / 500 [ 30%] (Warmup)
## Chain 1: Iteration: 200 / 500 [ 40%] (Warmup)
## Chain 1: Iteration: 250 / 500 [ 50%] (Warmup)
## Chain 1: Iteration: 251 / 500 [ 50%] (Sampling)
## Chain 1: Iteration: 300 / 500 [ 60%] (Sampling)
## Chain 1: Iteration: 350 / 500 [ 70%] (Sampling)
## Chain 1: Iteration: 400 / 500 [ 80%] (Sampling)
## Chain 1: Iteration: 450 / 500 [ 90%] (Sampling)
## Chain 1: Iteration: 500 / 500 [100%] (Sampling)
## Chain 1:
## Chain 1: Elapsed Time: 0.631 seconds (Warm-up)
## Chain 1: 0.404 seconds (Sampling)
## Chain 1: 1.035 seconds (Total)
## Chain 1:## mean se 5% 95% ess Rhat
## mu0 -0.0196 0.0010 -0.0459 0.0062 270.6255 0.9974
## sigma0 0.0840 0.0010 0.0632 0.1131 223.3145 1.0020
## arch 0.4803 0.0036 0.3869 0.5794 268.0763 0.9975
## garch 0.3643 0.0035 0.2686 0.4504 220.0485 0.9982
## loglik -869.9713 0.0952 -872.6722 -868.2944 258.2028 0.9997
 #### Diagnóstico
#### Diagnóstico
Ao ajustar um modelo bayesiano usando bayesforecast (que usa internamente Stan), o resumo do objeto ajustado inclui estatísticas de diagnóstico para cada parâmetro estimado. Dois dos principais são ess e Rhat.
ess: Effective Sample Size
Definição: O tamanho efetivo da amostra (ESS) mede quanta informação independente suas cadeias MCMC contêm para um parâmetro.
Intuição: Mesmo que você extraia, digamos, 2000 amostras de cada cadeia, as amostras são autocorrelacionadas (não independentes). O ESS informa a quantas amostras independentes suas cadeias são efetivamente equivalentes.
Interpretação: - ESS mais alto → estimativas posterioris mais confiáveis. - Como regra geral: - ESS > 1000 por parâmetro → bom. - ESS < 100 → indica forte autocorrelação; considere executar cadeias mais longas ou melhorar a amostragem.
bayesforecast relata ambos:
- ess_bulk: para a parte central (volume) da posteriori.
- ess_tail: para as caudas (valores extremos) da posteriori.
—
11.12.2.2.1 Rhat: Fator Potencial de Redução de Escala
Definição:
Rhat compara a variância entre cadeias com a variância dentro da cadeia para cada parâmetro. É conhecido como estatística de Gelman-Rubin.
Intuição:
Se múltiplas cadeias se misturaram bem e convergiram para o mesmo posterior, suas variâncias devem ser semelhantes → Rhat ≈ 1.
Interpretação:
- Rhat ≈ 1.00 → boa convergência.
- Rhat > 1.00 → possível não convergência, indicam que você deve executar novamente ou reespecificar o modelo.
—
Exercício 11.32
- No Exemplo 11.54 verifique a equivalência entre os modelos ARCH\((2)\), GARCH\((0,2)\) de
tseriese GARCH\((2,0,0)\) debayesforecast. - Veja https://asael697.r-universe.dev/bayesforecast.
- Veja https://cran.r-universe.dev/bayesforecast/doc/manual.html.
- Veja http://fmwww.bc.edu/EC-C/S2014/823/EC823.S2014.nn09.slides.pdf.
11.12.3 IGARCH
R. F. Engle and Bollerslev (1986) propõem o modelo IGARCH, “uma nova classe de modelos definidos para serem integrados em variância” a partir da restrição \(\sum_{i=1}^{s} \alpha_i + \sum_{i=1}^{r} \beta_i = 1\) na Eq. (11.40).
11.12.4 EGARCH
Nelson (1991) propõe o modelo EGARCH com o objetivo de “acomodar a relação assimétrica entre os retornos das ações e as mudanças na volatilidade”. Seguindo a notação de Vrontos, Dellaportas, and Politis (2000),
\[\begin{equation} \log h_t = \alpha_0 + \sum_{i=1}^s \theta_i a_{t-i} + \gamma_i (|a_{t-i}|-E|a_{t-i}|) + \sum_{i=1}^r \beta_i h_{t-i} \tag{11.42} \end{equation}\]
\[\begin{equation} E|a_{t-i}| = \frac{\Gamma(\frac{2}{\nu})}{[\Gamma(\frac{1}{\nu}) \Gamma(\frac{3}{\nu})]^{1/2}} \tag{11.43} \end{equation}\]
11.12.5 ARMA-GARCH
Exemplo 11.57 Considere a série temporal r unlist(tsdl::meta_tsdl[20,2][[1]][[1]]) apresentada por r tsdl::meta_tsdl[20,1], disponível em tsdl::tsdl[[20]]22.
library(forecast)
library(bayesforecast)
# ajustando um modelo ARIMA automático
y <- tsdl::tsdl[[20]]
# ARIMA(2,1,2)
forecast::auto.arima(y)## Series: y
## ARIMA(2,1,2)
##
## Coefficients:
## ar1 ar2 ma1 ma2
## 1.3467 -0.3963 -1.7710 0.8103
## s.e. 0.0303 0.0287 0.0205 0.0194
##
## sigma^2 = 243.8: log likelihood = -11745.5
## AIC=23500.99 AICc=23501.01 BIC=23530.71# ajustando ARMA(2,2)-GARCH(1,1) para a série com uma diferença via bayesforecast
fit_garch_bayes <- bayesforecast::garch(y, order = c(1,1,0), arma = c(2,2))
fit_garch_varstan <- bayesforecast::varstan(fit_garch_bayes, iter = 500, chains = 1)##
## SAMPLING FOR MODEL 'tgarch' NOW (CHAIN 1).
## Chain 1: Rejecting initial value:
## Chain 1: Log probability evaluates to log(0), i.e. negative infinity.
## Chain 1: Stan can't start sampling from this initial value.
## Chain 1:
## Chain 1: Gradient evaluation took 0.000624 seconds
## Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 6.24 seconds.
## Chain 1: Adjust your expectations accordingly!
## Chain 1:
## Chain 1:
## Chain 1: Iteration: 1 / 500 [ 0%] (Warmup)
## Chain 1: Iteration: 50 / 500 [ 10%] (Warmup)
## Chain 1: Iteration: 100 / 500 [ 20%] (Warmup)
## Chain 1: Iteration: 150 / 500 [ 30%] (Warmup)
## Chain 1: Iteration: 200 / 500 [ 40%] (Warmup)
## Chain 1: Iteration: 250 / 500 [ 50%] (Warmup)
## Chain 1: Iteration: 251 / 500 [ 50%] (Sampling)
## Chain 1: Iteration: 300 / 500 [ 60%] (Sampling)
## Chain 1: Iteration: 350 / 500 [ 70%] (Sampling)
## Chain 1: Iteration: 400 / 500 [ 80%] (Sampling)
## Chain 1: Iteration: 450 / 500 [ 90%] (Sampling)
## Chain 1: Iteration: 500 / 500 [100%] (Sampling)
## Chain 1:
## Chain 1: Elapsed Time: 20.972 seconds (Warm-up)
## Chain 1: 36.134 seconds (Sampling)
## Chain 1: 57.106 seconds (Total)
## Chain 1: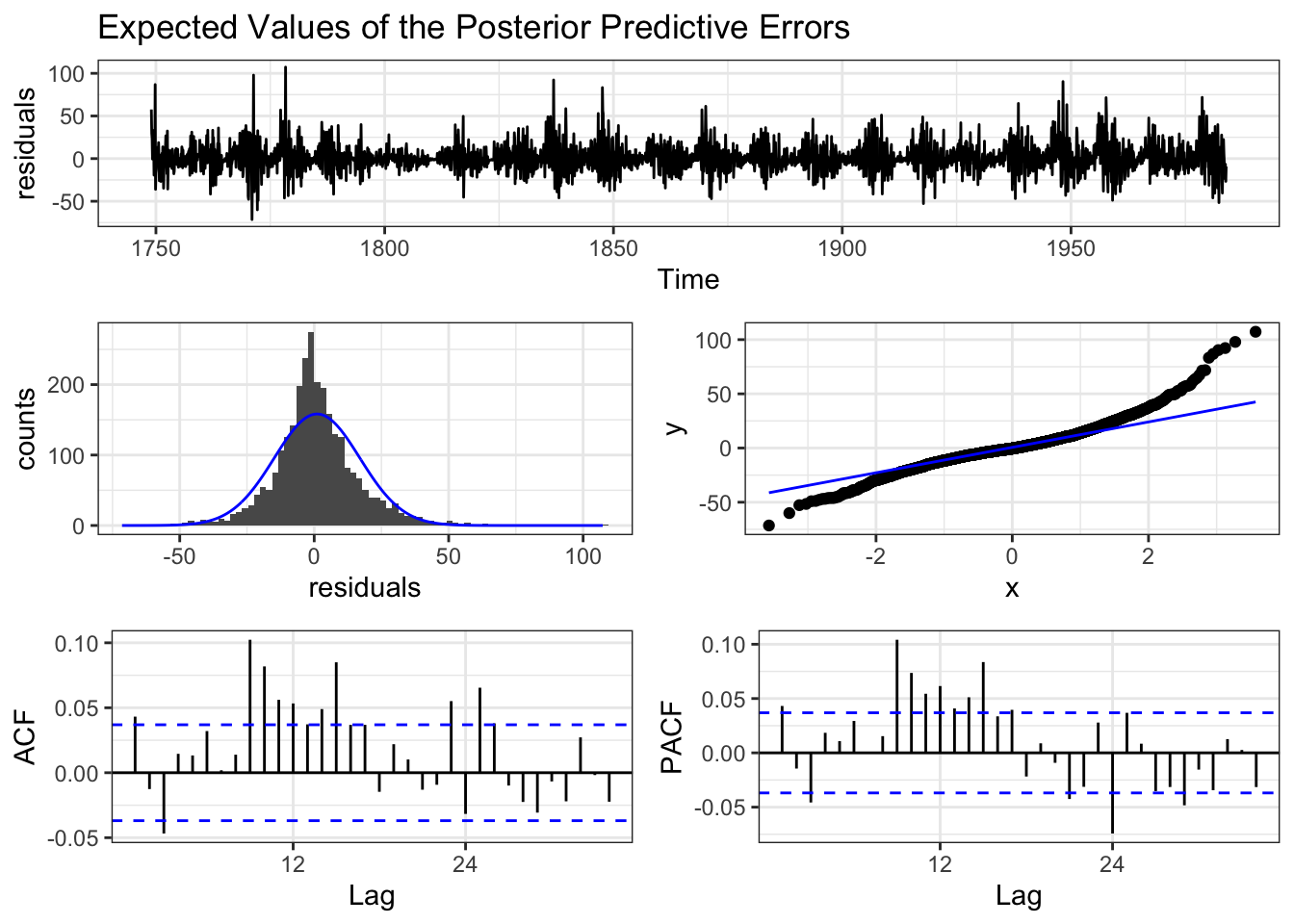
# # tomando uma diferença da série
# ydiff <- diff(y)
# fit_garch_bayes_diff <- bayesforecast::garch(ydiff, order = c(1,1,0), arma = c(2,2)) # ajusta ARMA(2,2)-GARCH(1,1)
# fit_garch_varstan_diff <- bayesforecast::varstan(fit_garch_bayes_diff, iter = 500, chains = 1)
# check_residuals(fit_garch_varstan_diff) # análise de resíduos
# bf <- bayesforecast::forecast(fit_garch_varstan_diff, h = 48)
# # desfazendo as diferenças
# bf$x <- diffinv(bf$x, xi = y[1])
# bf$mean <- diffinv(bf$mean, xi = y[length(y)])
# autoplot(bf) # projetando 48 passosA função rugarch::ugarchspec() (Galanos 2025) permite ajustar um modelo ARFIMA-GARCH, com variantes do modelo GARCH e variáveis externas (veja documentação).
# libs
library(forecast)
library(rugarch)
# ajustando um modelo ARIMA automático
y <- tsdl::tsdl[[37]]
# ARIMA(1,1,1)
(fit_arima <- forecast::auto.arima(y))## Series: y
## ARIMA(1,1,1)
##
## Coefficients:
## ar1 ma1
## -0.5322 0.9476
## s.e. 0.1389 0.0657
##
## sigma^2 = 95.43: log likelihood = -199.28
## AIC=404.57 AICc=405.05 BIC=410.54# avaliando a série ao quadrado e os resíduos ao quadrado do ARIMA(1,1,1)
par(mfrow=c(2,2))
acf(y^2)
pacf(y^2)
acf(fit_arima$residuals^2)
pacf(fit_arima$residuals^2)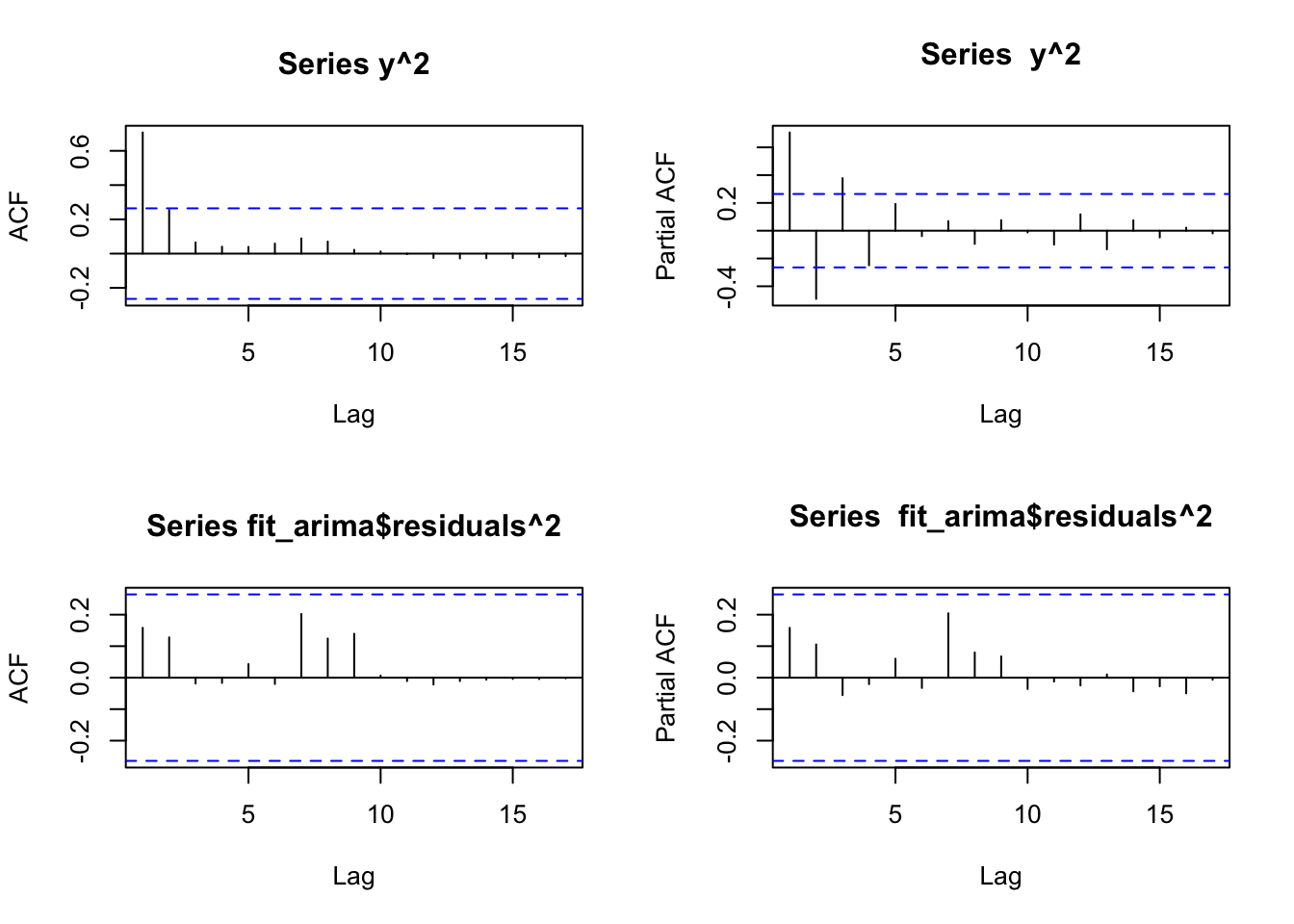
# tomando uma diferença da série
ydiff <- diff(y)
# ajustando ARMA-GARCH para a série com uma diferença via rugarch
spec <- ugarchspec(
mean.model = list(armaOrder = c(2,2), arfima = FALSE),
variance.model = list(garchOrder = c(0,1))
)
fit_garch <- ugarchfit(spec = spec, data = y, solver.control = list(trace=0))
fit_garch##
## *---------------------------------*
## * GARCH Model Fit *
## *---------------------------------*
##
## Conditional Variance Dynamics
## -----------------------------------
## GARCH Model : sGARCH(0,1)
## Mean Model : ARFIMA(2,0,2)
## Distribution : norm
##
## Optimal Parameters
## ------------------------------------
## Estimate Std. Error t value Pr(>|t|)
## mu 9.766934 0.729335 13.39155 0.00000
## ar1 0.973898 0.396209 2.45804 0.01397
## ar2 0.315425 0.525091 0.60071 0.54804
## ma1 -0.033162 0.048408 -0.68504 0.49332
## ma2 -1.091585 0.082780 -13.18655 0.00000
## omega 0.744145 5.849780 0.12721 0.89877
## beta1 0.999000 0.081919 12.19498 0.00000
##
## Robust Standard Errors:
## Estimate Std. Error t value Pr(>|t|)
## mu 9.766934 1.68699 5.789559 0.000000
## ar1 0.973898 1.37181 0.709935 0.477744
## ar2 0.315425 1.83388 0.171999 0.863438
## ma1 -0.033162 0.15540 -0.213397 0.831018
## ma2 -1.091585 0.29944 -3.645414 0.000267
## omega 0.744145 22.98703 0.032372 0.974175
## beta1 0.999000 0.32620 3.062507 0.002195
##
## LogLikelihood : -190.7872
##
## Information Criteria
## ------------------------------------
##
## Akaike 7.1923
## Bayes 7.4477
## Shibata 7.1645
## Hannan-Quinn 7.2911
##
## Weighted Ljung-Box Test on Standardized Residuals
## ------------------------------------
## statistic p-value
## Lag[1] 0.5043 0.4776
## Lag[2*(p+q)+(p+q)-1][11] 4.4612 0.9972
## Lag[4*(p+q)+(p+q)-1][19] 8.3802 0.7387
## d.o.f=4
## H0 : No serial correlation
##
## Weighted Ljung-Box Test on Standardized Squared Residuals
## ------------------------------------
## statistic p-value
## Lag[1] 6.099 0.01352
## Lag[2*(p+q)+(p+q)-1][2] 7.204 0.01052
## Lag[4*(p+q)+(p+q)-1][5] 9.123 0.01541
## d.o.f=1
##
## Weighted ARCH LM Tests
## ------------------------------------
## Statistic Shape Scale P-Value
## ARCH Lag[2] 2.055 0.500 2.000 0.1517
## ARCH Lag[4] 3.418 1.397 1.611 0.2099
## ARCH Lag[6] 3.773 2.222 1.500 0.3403
##
## Nyblom stability test
## ------------------------------------
## Joint Statistic: 13.3381
## Individual Statistics:
## mu 0.03699
## ar1 0.03213
## ar2 0.03141
## ma1 0.03472
## ma2 0.06054
## omega 0.58637
## beta1 0.46954
##
## Asymptotic Critical Values (10% 5% 1%)
## Joint Statistic: 1.69 1.9 2.35
## Individual Statistic: 0.35 0.47 0.75
##
## Sign Bias Test
## ------------------------------------
## t-value prob sig
## Sign Bias 0.06206 0.950761
## Negative Sign Bias 3.02333 0.003938 ***
## Positive Sign Bias 1.64253 0.106754
## Joint Effect 14.34129 0.002476 ***
##
##
## Adjusted Pearson Goodness-of-Fit Test:
## ------------------------------------
## group statistic p-value(g-1)
## 1 20 17.36 0.5652
## 2 30 25.18 0.6688
## 3 40 30.09 0.8463
## 4 50 44.09 0.6720
##
##
## Elapsed time : 0.09654498# previsão com o modelo
fcast <- ugarchforecast(fit_garch)
# plot(fcast, ask = FALSE)
# desfazendo a diferença
# fcast2 <- fcast
# fcast2@model$modeldata$data <- as.matrix(diffinv(fcast2@model$modeldata$data, xi = y[1]))
# fcast2@model$modeldata$data <- y[-1]
# fcast2@forecast$seriesFor <- as.matrix(diffinv(fcast2@forecast$seriesFor)[-1])
# plot(fcast2)